TEXT 文本标签
除了通过色块和连线来表示基因数据的关系,有时候我们也需要添加文字标签。 Text模块是plot模块中的子模块,是一种二维表示。一种基本的设置方法如下:
<plot>
type = text
file = data/6/genes.labels.txt
r1 = 0.8r
r0 = 0.6r
...
</plot>
你还需要为这个track设置好字体和标签大小。你还可以使用padding和rpadding去定义text的边缘:
label_font = light
label_size = 12p
rpadding = 5p
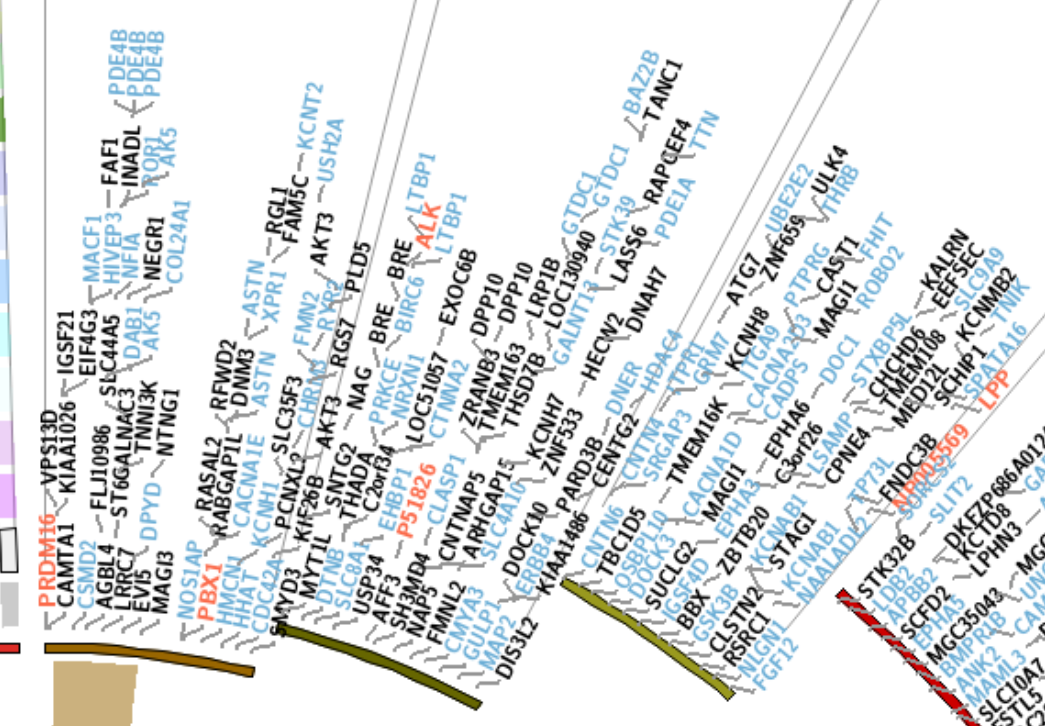
其基本数据格式除了要求染色体名称和起始终止位置外,还应有value一栏,即text的内容。下面例子使用了人类基因组中部分omim和cancer相关基因名称的数据。值得注意的是,通常在大量数据情况下,text不能完整显示,特别是邻近位置,所以对于text排版的设置尤为重要:
官方conf
<plot>
42 show = conf(show_text)
43 type = text
44 file = data/genes.txt #文本格式见下方
45 color = black
46 r1 = 0.73r
47 r0 = 0.415r
48 label_size = 9p
49 label_font = condensed
50 rpadding = 0r #径向填充
51 padding = 0r #角填充,具体细节见第二章
52
53 label_snuggle = yes
54 max_snuggle_distance = 1r
55 snuggle_sampling = 1
56 snuggle_tolerance = 0.25r
57
58 show_links = yes
59 link_dims = 4p,2p,5p,2p,2p
60 link_thickness = 2p
61 link_color = grey
62
63 <rules>
64 <rule>
65 condition = var(value) =~ /omim/ #正则匹配颜色区分
66 color = blue
67 flow = continue
68 </rule>
69 <rule>
70 condition = var(value) =~ /cancer/ #cancer就是红字
71 color = red
72 label_font = bold
73 flow = continue
74 </rule>
75 <rule>
76 # remove trailing _.* from label
77 condition = 1
78 value = eval(replace(var(value),qr/_.*/,""))
79 </rule>
80 </rules>
81
82 </plot>
genes.txt
1 hs10 100208866 100985609 HPSE2_other
2 hs10 105343775 105605154 SH3PXD2A_other
3 hs10 106390848 107014983 SORCS3_omim
4 hs10 108323411 108914282 SORCS1_omim
5 hs10 11087264 11418678 CUGBP2_other
6 hs10 11099898 11418678 CUGBP2_other
7 hs10 114196745 114568491 VTI1A_other
8 hs10 116843113 117698484 ATRNL1_other
9 hs10 1218073 1769670 ADARB2_omim
10 hs10 123738678 124004045 TACC2_omim
11 hs10 12431588 12908141 CAMK1D_other
12 hs10 127693414 128067055 ADAM12_omim
13 hs10 127719903 128067055 ADAM12_omim
14 hs10 128584012 129140769 DOCK1_omim
15 hs10 131155455 131455356 MGMT_omim
16 hs10 13725712 14412872 FRMD4A_other
17 hs10 14600564 14856902 FAM107B_other
18 hs10 16905970 17211822 CUBN_omim
19 hs10 18469611 18870694 CACNB2_omim